Target genes and specific reference genes, a unique marriage for successful RT-PCR experiments
Real-time PCR is the method of choice to measure the expression level of certain genes. But, these measurements require reference genes, also called housekeeping genes, as well as statistical analyses based on established mathematical methods. The identification of trusted reference genes is necessary and crucial for a successful and reproducible experiment.
But, a reference gene is not universal!
For instance, reference genes such as actin, ubiquitin or ribosomal genes are thought to be universally required for basic cellular functions and are constitutively and stably expressed under varying physiological and experimental conditions. But, recent works have revealed that the expression levels of these reference genes may vary depending on the cell type and experimental conditions.
The selection of a reference gene is based on the stability of its expression in all the experimental conditions studied. This stability of expression must therefore be verified for each new programmed experiment involving new conditions. Identifying such a gene is not easy and it is necessary to test several candidate genes to finally obtain this key element.
The expression stability of the reference genes is determined using the M-Value method of Vandesompele (Hellemans J et al. Genome Bio 2007). In this case, the measure of gene stability, the M-Value, is defined as the standard deviation of the logarithmically transformed expression values of the compared genes. This stability index and its decreasing value give rise to the rank and stability of the gene. In the first stage, the algorithm selects a pair of genes with the lowest M-Value, and the remaining genes are ranked according to the highest degree of compatibility with each other and with a geometric mean of the first pair. The evaluation is carried out on the basis of Ct values and for them is calculated: the geometric mean, the arithmetic mean, the minimum and maximum value, the standard deviation and the coefficient of variation.
In addition, an increasing number of institutions and journals ask scientists to present their results and the qPCR analyses using the MIQE (Minimum Information necessary for evaluating qPCR Experiments) guidelines, which require at least two reference genes.
Exploring MaRS for identify and select candidate reference genes
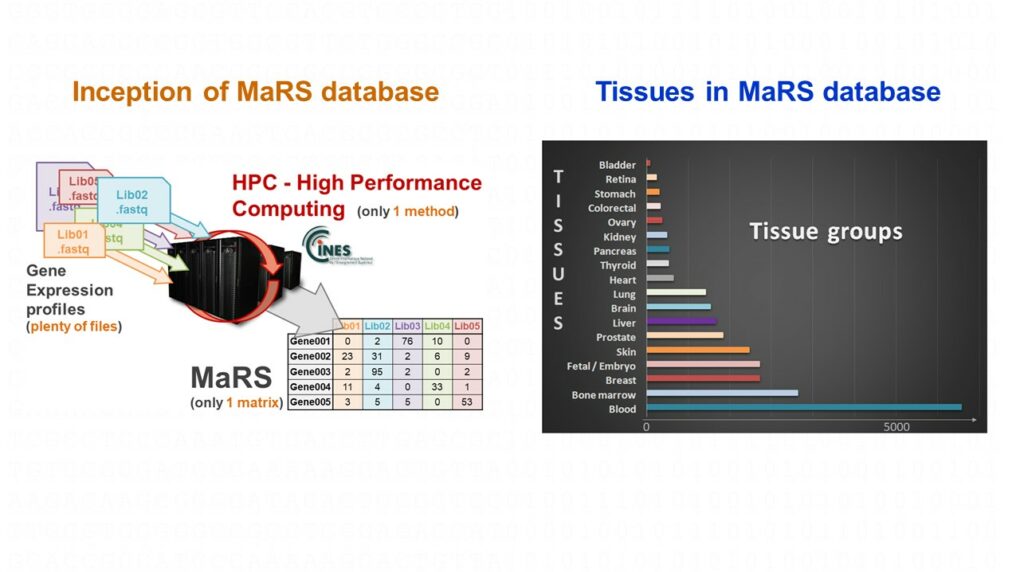
For several years, ACOBIOM identifies and selects reference genes to perform qPCR experiments for its clients, and to develop its own PCR diagnostics focused on precision medicine applications.
Thus, in a pre-discovery phase, ACOBIOM determines the most relevant reference genes according to a tissue, a pathology, or a temporal evolution thanks to its unique database of human gene expression profiles called MaRS (27,000 RNAseq profiles computed with a single method). This step allows the selection of between eight and twelve candidate genes depending on the problem. Then, in a second phase, ACOBIOM transfers these reference candidate genes to a real-time PCR platform to be validated. These two steps allow the identification and the validation of robust and precise reference genes associated with the selected target genes.
So, if the target genes, often selected after numerous analyses and studies, are considered as the Kings of an RT-PCR experiment, the reference genes are the Queens of this equation!
To make your qPCR experiment a success, ask ACOBIOM to organize a unique marriage between target genes and reference genes!

