RNA/DNA analyses to identify or confirm biomarkers and clinical endpoints
In order to prescribe the most adapted treatment in cancer, a range of diagnostics are performed to determine the presence of the pathology, its nature, its form or its classification. Thus, specific therapies are dedicated to particular sub-classes of cancer. This precision medicine approach exists mainly to treat breast cancers, and it has led to a significant improvement in the 10-year survival rate for this cancer: from 40% in 1971 to more than 80% in 2021.
Nevertheless, resistance or insensitivity to treatment remains an important issue. This is due to the lack of predictive markers for treatment resistance or to the lack of biomarkers associated with the monitoring of patients during treatment.
OncoSNIPE® a collaborative research program on the identification of biomarkers in oncology
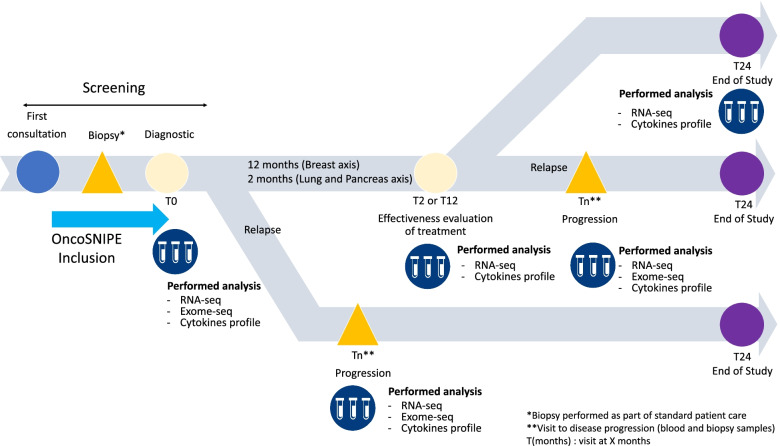
As part of the OncoSNIPE® collaborative research program, certain clinical endpoints have been associated to sampling (initial, follow-up, disease progression) and, after certain analyses, with specific biomarkers. Analyses will be performed in each cohort (cancer) independently. The objective is to identify early (baseline, predictive) or late (follow-up, monitoring) biomarkers associated with treatment resistance for each clinical endpoint.
Resistance is defined as a tumoral nonresponse according to the RECIST (Response Evaluation Criteria In Solid Tumor) criteria or absence of Pathologic Complete Response (pCR).
This study protocol led to the development of an optimized and standardized pipeline of bioinformatics and biostatistical tools to perform routine DNA and RNA analyses of the sequencing data.
The objective of exome/DNA analyses is the identification of putative DNA mutations, like InDels.
In the case of RNA-Seq analyses, different steps (normalization, mapping, counting) are necessary before the analysis of the differentially expressed genes, or the identification of RNA modifications like splicing or RNA editing. These analyses will lead to the identification or selection of putative RNA markers according to predetermined criteria and objectives: clustering, or exploratory research to highlight already known metabolic or signaling pathways.
The whole study protocol applied in this collaborative research program has been detailed in a recent publication: “OncoSNIPE® Study Protocol, a study of molecular profiles associated with development of resistance in solid cancer patients” (Vachenc et al, BMC Cancer 2022).
Acobiom uses its expertise to develop targeted therapies

As part of a collaborative project like OncoSNIPE®, or as part of a clinical trial evaluating the efficacy of a new therapy, ACOBIOM is convinced that the search and identification of omics biomarkers in peripheral blood is a relevant strategy to identify and select a sub-population of patients, responder to a specific therapy in a long term efficacy manner.
Thus, the identification of predictive biomarkers will make it possible to improve the results of current treatments, to reduce their side effects, and to identify the development of resistance.
This approach that may follow clinical end-points could also allow the identification of prognostic biomarkers associated with new therapies.
Indeed, ACOBIOM can exploit blood samples and associated data, collected during clinical trials, to optimize the development of targeted therapies or immunotherapies in predefined patient subgroups and thus, de-risk new projects and maximize the chances of success of clinical trials.
In this case, the company uses its proprietary omics-based machine-learning approach to determine different patient sub-populations, to identify the “good” and “bad” responders, and to set up a blood gene signature score to prognosticate patient responses and to pave the way for treatment personalization.
This predictive enrichment strategy reduces variability in the studied patient population and allows to select patients that are most likely to respond to a specific drug than other patients with the condition being treated.

